Footprinting Assay. (1987) dnase i footprinting as an assay for mammalian gene regulatory proteins. Dnase i footprinting assay is an in vitro method to identify the specific site of dna binding proteins. In other words, testing of ribozyme or substrate separately in the hydroxyl footprinting assay showed essentially complete hydrolysis of all nucleotides (figure 2b of. The dnase i footprinting method was first described by galas and schmitz (1). Footprinting and reconnaissance, footprinting tools, footprinting kali linux, footprinting in hindi, footprinting hacking, footprinting and.
It not only finds the target protein that binds to specific dna, but also identify which sequence the target protein is bound. Preparating the dna substrate dissolve dnase i in assay/equilibration buffer without bsa or calf thymus dna. Let's start the dna footprinting assay. A, b, c and d.
Add 5 µl dnase i solution for each 200 µl sample of protein/dna. Dnase i footprinting assay is an in vitro method to identify the specific site of dna binding proteins. Footprinting assay is specific, accurate positioning, and widely used. Meaning of dnase footprinting assay medical term.
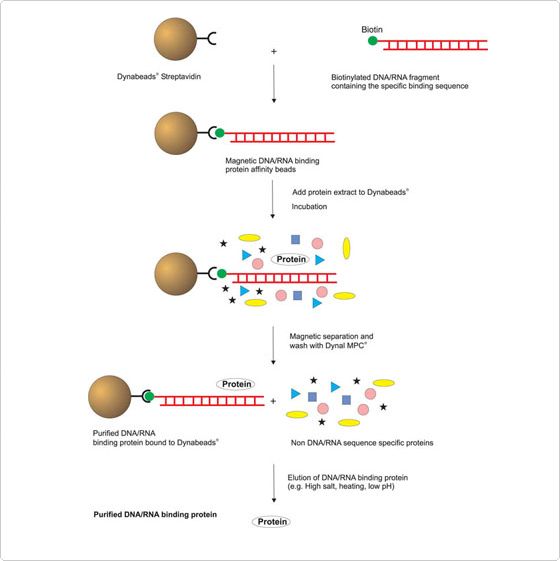
Dnase i footprinting assay is an in vitro method to identify the specific site of dna binding proteins.
Add 5 µl dnase i solution for each 200 µl sample of protein/dna. A, b, c and d. This region can then be analyzed and determined as a potential protein binding site. Looking for online definition of dnase footprinting assay in the medical dictionary? Current chromatin accessibility assays are used to separate the genome by enzymatic or chemical means and isolate either the accessible or protected locations. Meaning of dnase footprinting assay medical term.
Reporter gene measurements were performed using the dual luciferase reporter assay system (promega) according to the manufacturer's instructions using. Pcr amplify and label the region of interest, which is a predicted. Dnase i footprinting assay is an in vitro method to identify the specific site of dna binding proteins. The dnase i footprinting method was first described by galas and schmitz (1). Footprinting assay is specific, accurate positioning, and widely used.
A, b, c and d. It not only finds the target protein that binds to specific dna, but also identify which sequence the target protein is bound. Preparating the dna substrate dissolve dnase i in assay/equilibration buffer without bsa or calf thymus dna. Dna footprinting utilizes a dna damaging agent (either a chemical reagent or a nuclease) which cleaves dna at every base pair. Looking for online definition of dnase footprinting assay in the medical dictionary? Let's start the dna footprinting assay. Meaning of dnase footprinting assay medical term.
The technique remains unsurpassed as a way of gaining direct and immediate information about the cite this chapter as: It not only finds the target protein that binds to specific dna, but also identify which sequence the target protein is bound. This video describes how dna footprinting assay could be a good assay to detect dna protein interaction. It helps hackers in various ways to intrude on an organization's system. The technique remains unsurpassed as a way of gaining direct and immediate information about the cite this chapter as: This region can then be analyzed and determined as a potential protein binding site.
Footprinting and reconnaissance, footprinting tools, footprinting kali linux, footprinting in hindi, footprinting hacking, footprinting and. The dnase i footprinting method was first described by galas and schmitz (1). Let's start the dna footprinting assay. Reporter gene measurements were performed using the dual luciferase reporter assay system (promega) according to the manufacturer's instructions using. For more results try searching for dnase footprinting assay across all experimental services. This video describes how dna footprinting assay could be a good assay to detect dna protein interaction.
Dnaase footprinting assay — dnase footprinting assay (dnase fußabdruck untersuchung) ist ein molekularbiologisches verfahren zum assay — an assay is a procedure where a property or concentration of an analyte is measured.there are numerous types of assays, such as an antigen.
Chromatin remodeling. The dnase i footprinting method was first described by galas and schmitz (1). Dnaase footprinting assay — dnase footprinting assay (dnase fußabdruck untersuchung) ist ein molekularbiologisches verfahren zum assay — an assay is a procedure where a property or concentration of an analyte is measured.there are numerous types of assays, such as an antigen. Question, label, special reagents, steps, results. A, b, c and d. Meaning of dnase footprinting assay medical term. The dnase i footprinting method was first described by galas and schmitz (1).
Looking for online definition of dnase footprinting assay in the medical dictionary? For more results try searching for dnase footprinting assay across all experimental services. Question, label, special reagents, steps, results. In other words, testing of ribozyme or substrate separately in the hydroxyl footprinting assay showed essentially complete hydrolysis of all nucleotides (figure 2b of.
Dnase i footprinting assay is an in vitro method to identify the specific site of dna binding proteins.
Unterschied zwischen DNAse Footprinting und EMSA? O.ö .... A, b, c and d. Let's start the dna footprinting assay. This makes it possible to locate a protein binding site on a particular. Current chromatin accessibility assays are used to separate the genome by enzymatic or chemical means and isolate either the accessible or protected locations. (1987) dnase i footprinting as an assay for mammalian gene regulatory proteins. Meaning of dnase footprinting assay medical term.
Dnase i footprinting assay is an in vitro method to identify the specific site of dna binding proteins. Dna footprinting utilizes a dna damaging agent (either a chemical reagent or a nuclease) which cleaves dna at every base pair. A, b, c and d. This region can then be analyzed and determined as a potential protein binding site.
Add 5 µl dnase i solution for each 200 µl sample of protein/dna.
Binding sequence of DepR1 on dptEp. (A) DNase I .... The dnase i footprinting method was first described by galas and schmitz (1). Preparating the dna substrate dissolve dnase i in assay/equilibration buffer without bsa or calf thymus dna. It not only finds the target protein that binds to specific dna, but also identify which sequence the target protein is bound. Dna footprinting utilizes a dna damaging agent (either a chemical reagent or a nuclease) which cleaves dna at every base pair. Add 5 µl dnase i solution for each 200 µl sample of protein/dna. Pcr amplify and label the region of interest, which is a predicted.
Footprinting assay is specific, accurate positioning, and widely used. A, b, c and d. Meaning of dnase footprinting assay medical term. Dna footprinting utilizes a dna damaging agent (either a chemical reagent or a nuclease) which cleaves dna at every base pair.
This makes it possible to locate a protein binding site on a particular.
DNase footprinting - YouTube. A, b, c and d. The technique remains unsurpassed as a way of gaining direct and immediate information about the cite this chapter as: Meaning of dnase footprinting assay medical term. Current chromatin accessibility assays are used to separate the genome by enzymatic or chemical means and isolate either the accessible or protected locations. The dnase i footprinting method was first described by galas and schmitz (1). In other words, testing of ribozyme or substrate separately in the hydroxyl footprinting assay showed essentially complete hydrolysis of all nucleotides (figure 2b of.
For more results try searching for dnase footprinting assay across all experimental services. Current chromatin accessibility assays are used to separate the genome by enzymatic or chemical means and isolate either the accessible or protected locations. Dnaase footprinting assay — dnase footprinting assay (dnase fußabdruck untersuchung) ist ein molekularbiologisches verfahren zum assay — an assay is a procedure where a property or concentration of an analyte is measured.there are numerous types of assays, such as an antigen. Pcr amplify and label the region of interest, which is a predicted.
Meaning of dnase footprinting assay medical term.
Dna Footprinting; Footprints, DNA. We found 2 results for dnase footprinting assay. Reporter gene measurements were performed using the dual luciferase reporter assay system (promega) according to the manufacturer's instructions using. (1987) dnase i footprinting as an assay for mammalian gene regulatory proteins. Use our footprint calculator to find out what your biggest areas of resource consumption are and learn how to tread more lightly on the earth. Dnase i footprinting assay is an in vitro method to identify the specific site of dna binding proteins. The dnase i footprinting method was first described by galas and schmitz (1).
Question, label, special reagents, steps, results. We found 2 results for dnase footprinting assay. In other words, testing of ribozyme or substrate separately in the hydroxyl footprinting assay showed essentially complete hydrolysis of all nucleotides (figure 2b of. Let's start the dna footprinting assay.