Dnase 1 Footprinting. After a limited digestion with dnase i. Dnase i footprinting is used to precisely localise the position that a dna binding protein, e.g. A transcription factor, binds to a dna fragment. This technique can be used to study interactions between proteins and dna both. It is just a guide.
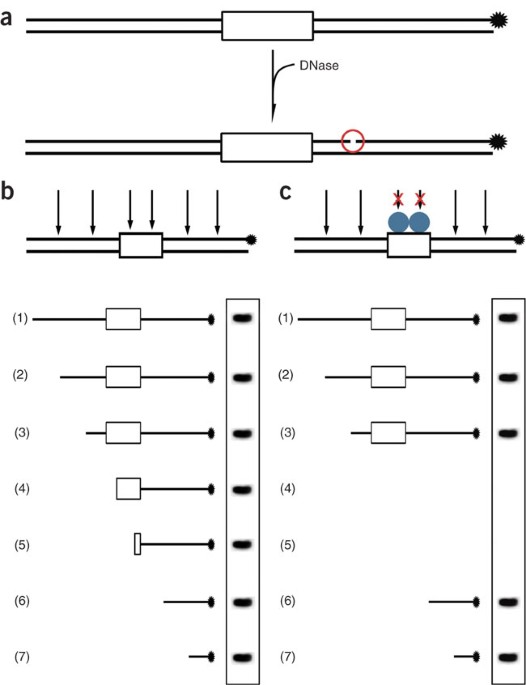
This is done in order to determine the footprint flanks. • finding a binding site • footprinting a binding site • affinity of interaction • crystallisation • structure solution. The following is a description for footprinting the tata complex. From gel electrophoresis 68 it has been deduced that the dna double helix is unwound.
This is a good concentration to start with. Lanes 1 and 4 had no protein lanes 2 and 3 had 2 different amounts of protein. From gel electrophoresis 68 it has been deduced that the dna double helix is unwound. Dnase i footprinting dms footprinting.
The cumulative skellam distribution function (package 'skellam') is used to detect significant normalized count differences of opposed sign at each dna strand.
Dnase i, or deoxyribonuclease i, is an endonuclease isolated from bovine pancreas. Many of the papers on identifying small regions protected from dnase i cleavage by a bound transcription factor do not seem to make their software available. The cumulative skellam distribution function (package 'skellam') is used to detect significant normalized count differences of opposed sign at each dna strand. Dnaase footprinting assay — dnase footprinting assay (dnase fußabdruck untersuchung) ist ein molekularbiologisches verfahren zum aufspüren von dna protein interaktionen. A method for determining the location of a protein binding site, dnase i footprinting analysis involves endonuclease treatment of an end labeled dna fragment bound to a protein. In the dnase i footprinting experiment shown here, a researcher began with a sample of cloned dna 300 bp in length.
This is a good concentration to start with. The cumulative skellam distribution function (package 'skellam') is used to detect significant normalized count differences of opposed sign at each dna strand. Dnase i footprinting is used to precisely localise the position that a dna binding protein, e.g. Dnase i, or deoxyribonuclease i, is an endonuclease isolated from bovine pancreas. Sample of a dnase i footprinting gel.
The cumulative skellam distribution function (package 'skellam') is used to detect significant normalized count differences of opposed sign at each dna strand. In the dnase i footprinting experiment shown here, a researcher began with a sample of cloned dna 300 bp in length. Lecture explains about dnase footprinting assay. On the other hand, it does not cut randomly as its activity is affected by local dna structure, thus giving an uneven ladder. The benefit of their training on specific motifs is that there appears to be a unique dnase cutting pattern for each tf. Dabei wird die tatsache ausgenutzt, dass dna an stellen, an denen ein protein gebunden ist, zu einem… … Dnase i footprinting assay is an in vitro method to identify the specific site of dna binding proteins.
You may find it necessary. This technique can be used to study interactions between proteins and dna both. It is just a guide. It assumes knowledge of the unerlying biological method, and is meant to help those that generally understand how to use the terminal to perform simple bioinformatic analyses. Lanes 1 and 4 had no protein lanes 2 and 3 had 2 different amounts of protein. Dabei wird die tatsache ausgenutzt, dass dna an stellen, an denen ein protein gebunden ist, zu einem… …
Many of the papers on identifying small regions protected from dnase i cleavage by a bound transcription factor do not seem to make their software available. Sample of a dnase i footprinting gel. It assumes knowledge of the unerlying biological method, and is meant to help those that generally understand how to use the terminal to perform simple bioinformatic analyses. Dnase i footprinting assay is an in vitro method to identify the specific site of dna binding proteins. It is just a guide. Optimized snakemake pipelines for atac and dnase1 footprinting are available for download at the.
The benefit of their training on specific motifs is that there appears to be a unique dnase cutting pattern for each tf.
DNase I footprinting analysis reveals a region of .... It assumes knowledge of the unerlying biological method, and is meant to help those that generally understand how to use the terminal to perform simple bioinformatic analyses. Dnase i, or deoxyribonuclease i, is an endonuclease isolated from bovine pancreas. Optimized snakemake pipelines for atac and dnase1 footprinting are available for download at the. The cumulative skellam distribution function (package 'skellam') is used to detect significant normalized count differences of opposed sign at each dna strand. This technique can be used to study interactions between proteins and dna both. Sample of a dnase i footprinting gel.
Limited digestion yields fragments terminating everywhere except in the footprint region, which is protected from digestion. Powerful and fairly rapid methods for mapping where and how proteins bind tightly to dna 2 ways: It assumes knowledge of the unerlying biological method, and is meant to help those that generally understand how to use the terminal to perform simple bioinformatic analyses. • finding a binding site • footprinting a binding site • affinity of interaction • crystallisation • structure solution.
Dnase i footprinting dms footprinting.
fig5:Four base recognition by triplex-forming .... The following is a description for footprinting the tata complex. It is just a guide. In the dnase i footprinting experiment shown here, a researcher began with a sample of cloned dna 300 bp in length. It assumes knowledge of the unerlying biological method, and is meant to help those that generally understand how to use the terminal to perform simple bioinformatic analyses. • finding a binding site • footprinting a binding site • affinity of interaction • crystallisation • structure solution. Different protein fractions may require 2 ul of dnase 1 solution with dnase 1 at 0.5 u/ul will be needed for each sample.
• finding a binding site • footprinting a binding site • affinity of interaction • crystallisation • structure solution. This technique can be used to study interactions between proteins and dna both. Dnaase footprinting assay — dnase footprinting assay (dnase fußabdruck untersuchung) ist ein molekularbiologisches verfahren zum aufspüren von dna protein interaktionen. • finding a binding site • footprinting a binding site • affinity of interaction • crystallisation • structure solution.
Dnase i footprinting is used to precisely localise the position that a dna binding protein, e.g.
Summary of DNase I footprinting with CI. The effects of CI .... Powerful and fairly rapid methods for mapping where and how proteins bind tightly to dna 2 ways: A method for determining the location of a protein binding site, dnase i footprinting analysis involves endonuclease treatment of an end labeled dna fragment bound to a protein. It not only finds the target protein that binds to specific dna, but also identify which sequence the target protein is bound. You may find it necessary. This is a good concentration to start with. Lecture explains about dnase footprinting assay.
It is just a guide. Dnase i footprinting is used to precisely localise the position that a dna binding protein, e.g. The following is a description for footprinting the tata complex. Powerful and fairly rapid methods for mapping where and how proteins bind tightly to dna 2 ways:
Dnase i footprinting is used to precisely localise the position that a dna binding protein, e.g.
DNase I footprinting analyses for BreR at the breAB .... Dnaase footprinting assay — dnase footprinting assay (dnase fußabdruck untersuchung) ist ein molekularbiologisches verfahren zum aufspüren von dna protein interaktionen. This is a good concentration to start with. Dnase i footprinting is used to precisely localise the position that a dna binding protein, e.g. A transcription factor, binds to a dna fragment. It assumes knowledge of the unerlying biological method, and is meant to help those that generally understand how to use the terminal to perform simple bioinformatic analyses. This dna contained a eukaryotic promoter for rna polymerase ii.
It is just a guide. This is done in order to determine the footprint flanks. Lanes 1 and 4 had no protein lanes 2 and 3 had 2 different amounts of protein. After a limited digestion with dnase i.
On the other hand, it does not cut randomly as its activity is affected by local dna structure, thus giving an uneven ladder.
DNAse footprinting assay - YouTube. Dabei wird die tatsache ausgenutzt, dass dna an stellen, an denen ein protein gebunden ist, zu einem… … Dabei wird die tatsache ausgenutzt, dass dna an stellen, an denen ein protein gebunden ist, zu einem… … Dnaase footprinting assay — dnase footprinting assay (dnase fußabdruck untersuchung) ist ein molekularbiologisches verfahren zum aufspüren von dna protein interaktionen. For the sample loaded in lane 1, no proteins were added. It not only finds the target protein that binds to specific dna, but also identify which sequence the target protein is bound. This is a good concentration to start with.
Dnase i, or deoxyribonuclease i, is an endonuclease isolated from bovine pancreas. A transcription factor, binds to a dna fragment. Dnaase footprinting assay — dnase footprinting assay (dnase fußabdruck untersuchung) ist ein molekularbiologisches verfahren zum aufspüren von dna protein interaktionen. This is a good concentration to start with.